Developing isoxazole as a native photo-cross-linker for photoaffinity labeling and chemoproteomics
 © 2022 Wiley-VCH GmbH
© 2022 Wiley-VCH GmbHAbstract
Photoaffinity labeling is a powerful technique to interrogate drug-protein interactions in native cellular environments. Photo-cross-linkers are instrumental for this technique. However, the introduction of unnatural photo-cross-linkers may significantly reduce the bioactivity of the drug, thus impairing the chemoproteomic outcomes. Herein, we developed a common pharmacophore, isoxazole, into a natively embedded photo-cross-linker for chemoproteomics, which minimally perturbs the drug structure. The photo-cross-linking reactions of the isoxazole were thoroughly investigated for the first time. Functionalized isoxazoles were then designed and applied to protein labeling, demonstrating the superior photo-cross-linking efficiency. Subsequently, two isoxazole-based drugs, Danazol and Luminespib, were employed in chemoproteomic studies, revealing their potential cellular targets. These results provide valuable strategies for future chemoproteomic study and drug development.
Highlights:
1. The photo-cross-linking of isoxazole towards biomolecules is thoroughly investigated first.
2. Functionalized isoxazole probes are designed for protein labeling, demonstrating superior photo-cross-linking efficiency.
3. Isoxazole-based drugs, Danazol and Luminespib, are employed in chemoproteomic studies, revealing their potential cellular targets.
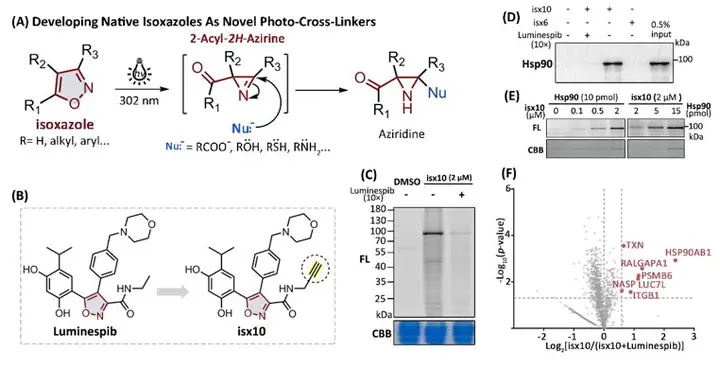
4. The probe derived from Luminespib (isx10) demonstrates superior specificity in labeling and imaging Hsp90.
5. Luminespib is investigated for covalent inhibition towards Hsp90 under UV irradiation.
Supplementary Information can be accessed here.